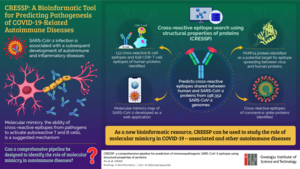
image: A new bioinformatics pipeline helps investigate the mechanism underlying the development of autoimmune diseases following SARS-CoV-2 infection
view more
Credit: Gwangju Institute of Science and Technology (GIST)
The SARS-CoV-2, or the novel coronavirus, has affected more than 500 million people worldwide. Apart from the symptoms associated with COVID-19 infection, it has recently been reported that the virus also leads to a subsequent development of autoimmune diseases in patients.
Autoimmune diseases like rheumatoid arthritis, lupus, or multi-inflammatory syndromes arise when the immune system confuses healthy cells with pathogens and starts attacking them. But, the precise mechanism underlying this “breach of self-tolerance” is unknown. One of the possible mechanisms suggested to be involved is what is called “molecular mimicry,” in which an autoimmune reaction is triggered when a T-cell receptor or an antibody produced from a B-cell directed against a specific antigen (foreign body) binds with an autoantigen, which is an antigen produced from our own body. This occurs due to a molecular or structural resemblance between the “epitopes” (the part of antigen attached to the antibody) of the antigens. However, a comprehensive investigation of the role of molecular mimicry in the development of such autoimmune diseases has not yet been performed due to the complexity of the epitope search and the lack of standardized tools.
To this end, a team of researchers from the Gwangju Institute of Science and Technology (GIST) led by Prof. Jihwan Park developed a new bioinformatics pipeline. Their new tool, called cross-reactive-epitope-search-using-structural-properties-of-proteins (CRESSP), was recently reported in the journal Briefings in Bioinformatics. “Previous studies on molecular mimicry used bioinformatics pipelines different from one another that often involved complex algorithms and were not scalable to proteome scales. In light of this, we developed a pipeline that is easily accessible and scalable,” explains Prof. Park. “It uses the structural properties of proteins to identify epitope similarities between two proteins of interest, such as human and SARS-CoV-2 proteins.”
Using CRESSP, the team screened 4,911,245 proteins from 196,352 SARS-CoV-2 genomes obtained from an open-access database. The pipeline narrowed down 133 cross-reactive B-cell and 648 CD8+ T-cell epitopes that could be responsible for COVID-related autoimmune diseases. It further identified a protein target, PARP14, to be a potential initiator of epitope spreading between COVID-19 virus and human lung proteins.
The pipeline also predicted the cross-reactive epitopes of different coronavirus spike proteins. Moreover, the team developed an interactive web application to enable an interactive visualization of the molecular mimicry map of SARS-CoV-2. The pipeline is also available as an open-source package.
The team hopes their new tool will facilitate comparison between studies, providing a robust framework for further investigation on molecular mimicry and autoimmune diseases. “Although autoimmune diseases affect less than 10% of the population, studying them is important since it severely impacts the quality of life. Our new tool can be used to study the possible involvement of molecular mimicry in the development of other autoimmune conditions in a systemic and scalable manner,” concludes Prof Park.
Hopefully, the new invention will help us deal with SARS-CoV-2 and other viral infections better.
***
Reference
DOI: https://doi.org/10.1093/bib/bbac056
Authors: Hyunsu An, Minho Eun, Jawoon Yi, Jihwan Park
Affiliations: School of Life Sciences, Gwangju Institute of Science and Technology (GIST)
About the Gwangju Institute of Science and Technology (GIST)
The Gwangju Institute of Science and Technology (GIST) was founded in 1993 by the Korean government as a research-oriented graduate school to help ensure Korea’s continued economic growth and prosperity by developing advanced science and technology with an emphasis on collaboration with the international community. Since that time, GIST has pioneered a highly regarded undergraduate science curriculum in 2010 that has become a model for other science universities in Korea. To learn more about GIST and its exciting opportunities for researchers and students alike, please visit: http://www.gist.ac.kr/.
About the author
Jihwan Park is an associate professor at the School of Life Sciences at GIST in Korea. He received a Ph.D. in epigenomics from POSTECH in Korea. He continued his study on genetics and epigenetics of human diseases using multi-omics analysis and single-cell analysis at the University of Pennsylvania in the USA under the supervision of Dr. Katalin Susztak. Currently, his group explores the molecular mechanisms of diseases such as cancer and chronic diseases using single-cell analysis. His lab also focuses on developing single-cell sequencing techniques for full-length mRNA sequencing and cell lineage tracing in cancer evolution and organogenesis.
Journal
Briefings in Bioinformatics
Method of Research
Computational simulation/modeling
Subject of Research
Not applicable
Article Title
CRESSP: a comprehensive pipeline for prediction of immunopathogenic SARS-CoV-2 epitopes using structural properties of proteins
Article Publication Date
28-Feb-2022
COI Statement
The authors declare no competing interests.
Disclaimer: AAAS and EurekAlert! are not responsible for the accuracy of news releases posted to EurekAlert! by contributing institutions or for the use of any information through the EurekAlert system.